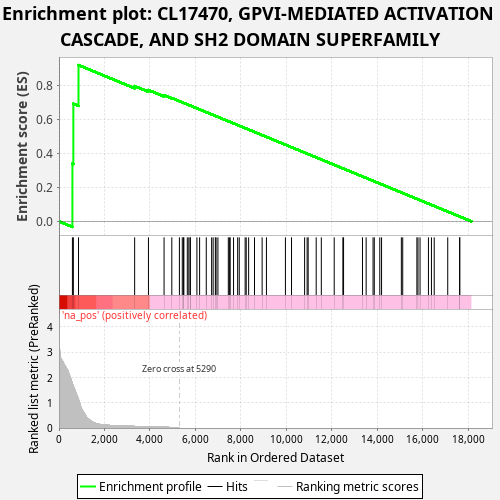
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Alopecia custom__Alopecia_custom-both_sexes- |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CL17470, GPVI-MEDIATED ACTIVATION CASCADE, AND SH2 DOMAIN SUPERFAMILY |
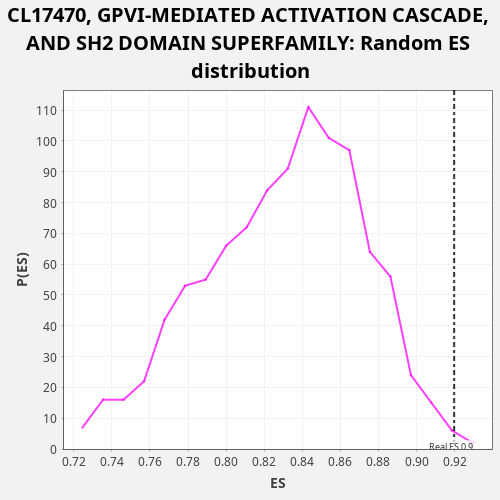
| Enrichment Score (ES) | 0.9197545 |
| Normalized Enrichment Score (NES) | 1.1077741 |
| Nominal p-value | 0.003 |
| FDR q-value | 0.3264449 |
| FWER p-Value | 0.836 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | GP6 | 590 | 1.736 | 0.3399 | Yes |
| 2 | SYK | 631 | 1.653 | 0.6922 | Yes |
| 3 | PIK3R6 | 858 | 1.119 | 0.9198 | Yes |
| 4 | INSRR | 3326 | 0.061 | 0.7962 | No |
| 5 | UBASH3A | 3933 | 0.048 | 0.7729 | No |
| 6 | VAV3 | 4619 | 0.035 | 0.7426 | No |
| 7 | GRB2 | 4960 | 0.010 | 0.7258 | No |
| 8 | FGR | 5294 | -0.000 | 0.7074 | No |
| 9 | CD79B | 5420 | -0.000 | 0.7005 | No |
| 10 | BTK | 5481 | -0.000 | 0.6972 | No |
| 11 | FYN | 5484 | -0.000 | 0.6971 | No |
| 12 | GAB2 | 5641 | -0.000 | 0.6884 | No |
| 13 | LCP2 | 5707 | -0.000 | 0.6848 | No |
| 14 | BCAR1 | 5768 | -0.000 | 0.6815 | No |
| 15 | PIK3CB | 5774 | -0.000 | 0.6812 | No |
| 16 | DAPP1 | 6063 | -0.000 | 0.6653 | No |
| 17 | TXK | 6182 | -0.000 | 0.6588 | No |
| 18 | LAT2 | 6475 | -0.000 | 0.6426 | No |
| 19 | BLNK | 6711 | -0.000 | 0.6296 | No |
| 20 | CRKL | 6777 | -0.000 | 0.6260 | No |
| 21 | GRAP2 | 6874 | -0.000 | 0.6207 | No |
| 22 | SOS2 | 6906 | -0.000 | 0.6190 | No |
| 23 | SLA2 | 6987 | -0.000 | 0.6146 | No |
| 24 | CD79A | 7444 | -0.000 | 0.5893 | No |
| 25 | PIK3R2 | 7491 | -0.000 | 0.5868 | No |
| 26 | PIK3CG | 7527 | -0.000 | 0.5848 | No |
| 27 | RAPGEF1 | 7674 | -0.000 | 0.5768 | No |
| 28 | GAB1 | 7855 | -0.000 | 0.5668 | No |
| 29 | CBL | 7925 | -0.000 | 0.5630 | No |
| 30 | ITK | 8184 | -0.000 | 0.5487 | No |
| 31 | PDGFRB | 8231 | -0.000 | 0.5462 | No |
| 32 | ZAP70 | 8338 | -0.000 | 0.5403 | No |
| 33 | PIK3R3 | 8595 | -0.000 | 0.5261 | No |
| 34 | PIK3CA | 8931 | -0.000 | 0.5076 | No |
| 35 | PLCG1 | 9120 | -0.000 | 0.4972 | No |
| 36 | IRS4 | 9955 | -0.000 | 0.4510 | No |
| 37 | TEC | 10223 | -0.000 | 0.4362 | No |
| 38 | IGF1R | 10798 | -0.000 | 0.4045 | No |
| 39 | PIK3R5 | 10909 | -0.000 | 0.3984 | No |
| 40 | VAV1 | 10955 | -0.000 | 0.3959 | No |
| 41 | PIK3R1 | 11308 | -0.000 | 0.3764 | No |
| 42 | SHC3 | 11533 | -0.000 | 0.3640 | No |
| 43 | PIK3AP1 | 12100 | -0.000 | 0.3327 | No |
| 44 | VAV2 | 12481 | -0.000 | 0.3117 | No |
| 45 | SHC1 | 12511 | -0.000 | 0.3101 | No |
| 46 | FRS2 | 13343 | -0.000 | 0.2641 | No |
| 47 | CRK | 13504 | -0.000 | 0.2552 | No |
| 48 | IRS1 | 13808 | -0.000 | 0.2385 | No |
| 49 | PTK2 | 13862 | -0.000 | 0.2355 | No |
| 50 | INSR | 14104 | -0.000 | 0.2222 | No |
| 51 | PIK3CD | 14182 | -0.000 | 0.2179 | No |
| 52 | PTPN11 | 15052 | -0.000 | 0.1698 | No |
| 53 | SOCS4 | 15112 | -0.000 | 0.1666 | No |
| 54 | SHC4 | 15735 | -0.000 | 0.1322 | No |
| 55 | IRS2 | 15783 | -0.000 | 0.1296 | No |
| 56 | BCR | 15874 | -0.000 | 0.1246 | No |
| 57 | PTPN1 | 16243 | -0.000 | 0.1042 | No |
| 58 | SRC | 16375 | -0.000 | 0.0970 | No |
| 59 | PLCG2 | 16497 | -0.000 | 0.0903 | No |
| 60 | LAT | 17091 | -0.000 | 0.0574 | No |
| 61 | LYN | 17613 | -0.000 | 0.0286 | No |
| 62 | INS | 17622 | -0.000 | 0.0282 | No |